Brief Communications
Abstract
QTLNetworkR is an R package that aims to provide a user-friendly and platform-independent tool to visualize quantitative trait loci (QTL) mapping results. The graphical functions of the QTLNetworkR are based upon lattice and grid packages, and the graphical user interface (GUI) of the QTLNetworkR is built upon RGtk2 and gWidgetsRGtk2 packages. Six functions are designed to help visualize marker interval, putative QTL, QTL-by-environment interactions, marker interval interactions, epistasis, and the predicted genetic architecture of complex traits. It is especially helpful in profiling results for multiple traits at multiple environments. The current version of QTLNetworkR is able to accept QTL mapping results from QTLNetwork, and it is ready for possible extensions to import results from some other QTL mapping software packages. In addition, we presented a QTL mapping result in rice (Oryza sativa) as an example to describe the features of QTLNetworkR.
QTL analysis for blood glucose was performed using Map Manager QTXb20, a software program that uses a maximum likelihood algorithm with interval mapping and simultaneous search and that permits.
Qtl Mapping Example
1. Introduction
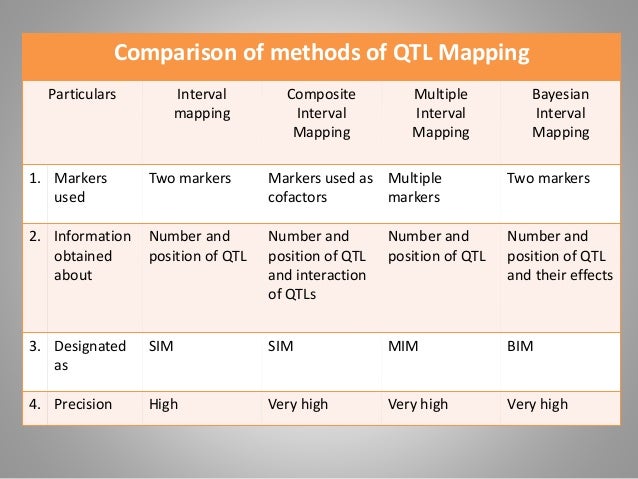
Mapping quantitative trait loci (QTL) is of great importance to understand the genetic architecture underlying complex traits (Mackay, ; Doebley, ), and many statistical methods have been proposed in the last two decades, such as interval mapping (Lander and Botstein, ; Haley and Knott, ), composite interval mapping (Zeng, ), and multiple interval mapping (Kao et al., ). Although QTL mapping packages such as MAPMAKER/QTL (Lander et al., ), QTLMapper (Wang et al., 1999), Map Manager (Manly et al., ), QTL Express (Seaton et al., ), R/qtl (Broman et al., ), MCQTL (Jourjon et al., ), WinQTLCart (Wang et al., 2006), and QGene 4.0 (Joehanes and Nelson, ) have been developed and released to public, none of them can map and visualize epistasis and QTL-by-environment (QE) interactions simultaneously.
Recently, a promising tool called QTLNetwork (Yang et al., ) has been developed based on a novel QTL mapping strategy (Yang et al., ) that can map single QTLs, epistasis, and QE interaction for experimental populations of various crosses. The command-line versions of QTLNetwork enable it run across most operating systems, such as Microsoft Windows, Mac OS, and Linux, but the graphical user interface (GUI) one is available for Windows only. Despite of its great visualization features, it remains drawbacks in the GUI version; e.g., it is merely able to illustrate one trait at a time and pale to present the QE interaction in detail. Moreover, the profiles from QTLNetwork are insufficient to prepare manuscripts as it lacks flexibility to change some primitives, such as fonts and scales of axis. In order to enhance the quality of the illustration for mapping results, we developed a user-friendly and platform-independent tool called QTLNetworkR to visualize QTL mapping results generated by QTLNetwork in a more convenient and informative way. Six functions were designed in QTLNetworkR for F-statistic profiles of marker interval analysis (looking for marker intervals potentially bound up with QTLs), one-dimensional (1D) genome scan for putative QTLs, QE interaction, and marker interval interaction analyses, two-dimensional (2D) genome scan for epistasis and the predicted genetic architecture.
2. Design and implementation
We designed and developed the QTLNetworkR as an add-on package in R (Ihaka and Gentleman, 1996) for its popularity in the academic community and platform-independence. To facilitate users, we provided a GUI for the QTLNetworkR based on RGtk2, an add-on R package for combining GUI toolkit GTK+ 2.0 with R, via the aid of another package gWidgetsRGtk2, both of which can be downloaded from the Comprehensive R Archive Network (CRAN, http://cran.r-project.org/). There are two alternative graphics systems in R, traditional graphics like S and grid, and the latter one, at least in this study, convinced us that it is a more flexible scaffold with adding further details for QTL mapping, e.g., epistasis and QE interactions. We consequently chose it as the basis of graphical functions in the QTLNetworkR.
The QTLNetworkR starts with a ‘file’ container for loading a QTL mapping result file (QTLNetwork output) and a linkage map file. In addition, it is embedded with a dialog notebook accommodating six tabbed pages, for functions designed, with options to control graphical output primitives, e.g., lines and text, to meet the requirement for preparing manuscripts in different formats.
Here, we present a QTL mapping result in rice (Oryza sativa) as an example to demonstrate the application of QTLNetworkR (unpublished data). It is a recombinant inbred line population on 10 morphological and yield-related traits from seven different environments. A total of 221 RFLP markers scored along the 12 rice chromosomes were used to construct the genetic linkage map and distributed over 15 linkage groups eventually.
The QTLNetworkR provides more enhanced visualization than the QTLNetwork (GUI version), including F-statistic profiles of 1D genome scan for the putative QTLs (Fig. (Fig.1a)1a) and 2D genome scan for the epistasis as well as a plot to summarize the network among QTL detected (Fig. (Fig.1b1b).
Visualization of QTL mapping results in QTLNetworkR for an inbred line population on multiple traits (unpublished data) in rice
(a) F-statistic profile of one-dimensional (1D) genome scan on linkage groups (LGs) 1 and 4 for grain rate (red), 1000 grain weight (blue), and full grain number (green), respectively. Linkage peaks that exceed the threshold F-value are considered as putative QTLs; (b) Predicted genetic architecture for grain rate (square), 1000 grain weight (triangle), and full grain number (circle). The loci with black fill represent epistatic QTLs without individual effects. QTLs with potentially pleiotropic effects are labeled by open rectangles. A: additive effect; AE: additive×environment interaction effect; AA: additive×additive epistatic effect; AAE: AA×environment interaction effect
- About R/qtl: About R; R/qtl is an extensible, interactive environment for mapping quantitative trait loci (QTL) in experimental crosses. It is implemented as an add-on package for the freely available and widely used statistical language/software R (see the R project homepage). The development of this software as an add-on to R allows us to take advantage of the basic mathematical and statistical functions,.
- TetraploidMap is a graphical user interface for calculating linkage maps for autotetraploid populations. It is suitable for handling markers scored on two parents and the full-sib offspring of a cross between them. TetraploidMap handles both codominant and dominant molecular markers, in all possible configurations, and takes into account the presence of null alleles in the analysis.
Furthermore, pleiotropy often occurs when a single locus influences multiple phenotypic traits, and many lines of evidence espoused its ubiquitousness (Wright, ; Clark et al., ). Growing experimental data are collected upon multiple traits and it would be of interest to discover QTLs of pleiotropic effects, if there is any. As the QTLNetwork is mute to profile multiple traits simultaneously, we offered an option to present multiple traits for 1D genome scan and the summarized QTL network to highlight the pleiotropic effects of QTLs (Figs. 1a and 1b). In this way, we may find that an epistatic QTL, without individual effect on one trait, could be a main-effect QTL for another trait, e.g., QTL 1-3 (Fig. (Fig.1b1b).
QE interaction is an essential nature for the genetic basis of complex trait. As such information has been limitedly presented in the QTLNetwork, we provided a more detailed illustration to show the main effect (A) across environments and the significant QTL effect (A+AE) in specific environments (Fig. (Fig.22).
Plot of the main QTL effects and the QTL-by-environment (QE) interaction effects for grain rate in rice
‘QTL’ represents the main effect of each putative QTL and ‘Q+QE1’ to ‘Q+QE7’ represent genetic effects (main QTL effects plus QE interaction effects) in 1 to 7 environments. The x-axis label, for example, ‘1–9’ represents a putative QTL locates at marker interval 9 on linkage group 1
The QTLNetworkR package is freely available at http://cran.r-project.org/web/packages/QTLNetworkR/index.html. Alternatively, it can be downloaded and installed directly in R environment with command: install.packages (pkgs=“QTLNetworkR”). Detailed information about QTLNetworkR can be looked up in its user manual from http://qtlnetworkr.r-forge.r-project.org/
3. Conclusions
QTLNetworkR provides a user-friendly and platform-independent tool to visualize QTL mapping results generated but not limited by QTLNetwork. Results from other QTL mapping packages, after reorganizing their formats, can also be imported and visualized. It is capable of presenting QTL mapping results for multiple traits and providing additional options to pronounce QE interaction. It can be easily run in R environment regardless of operating systems. As a QTL mapping visualization tool, the QTLNetworkR will be particularly helpful for profiling results for multiple traits at multiple environments.
Acknowledgments
We thank Dr. Guo-bo CHEN (Department of Agronomy, Zhejiang University, China) for his helpful comments.
Footnotes
*Project supported by the National Basic Research Program (973) of China (No. 2008CB117002), the Ministry of Agriculture Public Benefit Research Foundation of China (No. 200803034), and the National High-Tech R & D Program (863) of China (No. 2006AA10A102)